Abstract
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare and aggressive blood cancer and an ongoing therapeutic challenge. In a recent multiomics analysis, we characterized the unique molecular landscape of BPDCN. Beyond an expected classical pDC-derived subtype, we thereby identified a common(c)DC-enriched subtype, which was shown to predominantly arise in elderly patients, whilst harboring fewer mutations and an inferior outcome. The identification of several therapeutically targetable vulnerabilities may pave the way for precision oncology approaches toward this rare cancer. Several genomic alterations in both subtypes were found to be known determinants of the epigenetic regulation of gene expression. However, the epigenetic landscape of BPDCN remains sparsely characterized.
We therefore characterized the genome-wide DNA methylation patterns in 54 BPDCN patients on the Illumina MethylationEPIC BeadChip and correlated our findings with WES/RNA-seq data from both previous and new studies. Further, we evaluated our observations according to the newly defined subtypes of BPDCN.
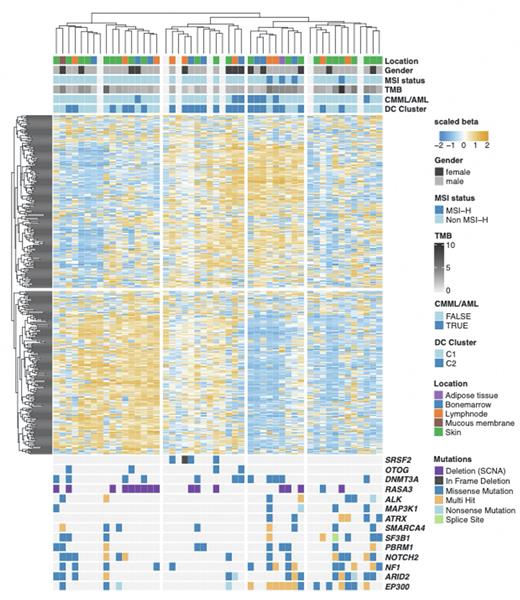
Unsupervised clustering of the 420 most variably methylated cytosine guanine dinucleotide (CpG) sites resulted in the segregation of the study cohort into four clusters. These were subsequently annotated genomically and transcriptionally in a clinical context. Cluster two was hereby shown to be of special interest as it was exclusively composed of C2-BPDCN cases with SRSF2 or DNMT3A mutations. Cluster four revealed a strong C1-BPDCN predominance and typical C1 driver mutations including EP300, ARID2 and NF1 (Figure 1).
Secondly, differential methylation analysis according to our recently established subtypes of BPDCN revealed a subset of significantly differentially methylated genes between the two clusters. Overall, we observed more methylation among C2 patients with regulation of several oncogenes and tumor suppressor genes such as NOTCH4 and MAP3K13 but a predominant regulation of inflammatory genes including TLR9, IL10RA and IL2RA.
Our current observations suggest that genome-wide DNA methylation levels resemble and potentially help shape clinically and genomically distinct subgroups of BPDCN. Synoptic analysis of the methylome in concert with gene-expression profiling data alongside ATAC-Seq of our BPDCN samples is currently under way in order to integratively assess chromatin accessibility and ultimately transcriptional impact.
Figure legend
Figure 1. Genome-wide DNA methylation profiling in blastic plasmacytoid dendritic cell neoplasm. Unsupervised clustering of the 420 most variably methylated CpG sites resulted in the segregation of the study cohort into four clusters. These were subsequently annotated genomically and transcriptionally in a clinical context.
Disclosures
Gebauer:Beigene: Other: Travel Support; Roche: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal